CAMP responsive element modulator
| CREM | |||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
 | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Identifiers | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Aliases | CREM, CREM-2, ICER, hCREM-2, cAMP responsive element modulator | ||||||||||||||||||||||||||||||||||||||||||||||||||
| External IDs | OMIM: 123812 MGI: 88495 HomoloGene: 84591 GeneCards: CREM | ||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
cAMP responsive element modulator is a protein that in humans is encoded by the CREM gene,[5][6][7] and it belongs to the cAMP-responsive element binding protein family. It has multiple isoforms, which act either as repressors or activators.[8] CREB family is important for in regulating transcription in response to various stresses, metabolic and developmental signals.[9] CREM transcription factors also play an important role in many physiological systems, such as cardiac function,[10] circadian rhythms,[11] locomotion and spermatogenesis.[12]
Function
This gene encodes a bZIP transcription factor that binds to the cAMP responsive element found in many viral and cellular promoters. It is an important component of cAMP-mediated signal transduction during the spermatogenetic cycle, as well as other complex processes. Alternative promoter and translation initiation site usage allows this gene to exert spatial and temporal specificity to cAMP responsiveness. Multiple alternatively spliced transcript variants encoding several different isoforms have been found for this gene, with some of them functioning as activators and some as repressors of transcription.[7]
Gene location
The chromosomal location of CREM gene is at 10p11.21, where it starts at 35415769 and ends at 35501886 bp from pter ( according to hg19-Feb_2009)[13]
Interactions
CAMP responsive element modulator has been shown to interact with FHL5.[14][15]
Disease relevance of CREM
Panic disorder
One study reported the DNA sequence variations in the gene for CREM in panic disorder patients. It showed a significant excess of the shorter eight-repeat allele and of genotypes containing the eight-repeat allele in panic disorder patients.[16] The observed associations were limited to panic disorder without agoraphobia, and they were more prominent in females. But, the independent Italian and Spanish samples in this study did not support their results. Another family-based study showed little evidence of any susceptibility locus for panic disorder either within the CREM gene or in a nearby region on chromosome 10p11[17]
Spermiogenesis deficiency
CREM has been shown to be a master-switch regulator in testis.[18] It plays an important role in the regulation of the expression of post-meiotic genes, and this has been supported by several studies using CREM-mutation mice.[19] The results showed the first step in the process of sperm formation would be blocked if the germ cell development in mice CREM gene were disrupted. The cAMP response element sites can be found in the promoter region of some postmeiotic genes, so that the CREM can target and regulate these genes.[18]
Two studies proved that treat the rats with Salvia hypoleuca and Alpina galanga can significantly increased the CREM gene expression.[20][21]
Systemic lupus erythematousus
Less IL-2 will be produced from T cells in humans or mice with systemic lupus erythematousus (SLE). Some studies showed that an increased level CREM was presented in the nucleus of T lymphocytes from SLE patients. The CREM bound to the -180 site of the IL-2 promoter to repress its transcription.[22]
References
- ^ a b c GRCh38: Ensembl release 89: ENSG00000095794 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000063889 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ Meyer TE, Habener JF (Nov 1992). "Cyclic AMP response element binding protein CREB and modulator protein CREM are products of distinct genes". Nucleic Acids Research. 20 (22): 6106. doi:10.1093/nar/20.22.6106. PMC 334485. PMID 1461747.
- ^ Masquilier D, Foulkes NS, Mattei MG, Sassone-Corsi P (Nov 1993). "Human CREM gene: evolutionary conservation, chromosomal localization, and inducibility of the transcript". Cell Growth & Differentiation. 4 (11): 931–7. PMID 7916662.
- ^ a b "Entrez Gene: CREM cAMP responsive element modulator".
- ^ Foulkes NS, Sassone-Corsi P (1992-02-07). "More is better: activators and repressors from the same gene". Cell. 68 (3): 411–414. doi:10.1016/0092-8674(92)90178-f. ISSN 0092-8674. PMID 1739963. S2CID 34290449.
- ^ Sassone-Corsi P (1995-01-01). "Transcription factors responsive to cAMP". Annual Review of Cell and Developmental Biology. 11: 355–377. doi:10.1146/annurev.cb.11.110195.002035. ISSN 1081-0706. PMID 8689562.
- ^ Isoda T, Paolocci N, Haghighi K, Wang C, Wang Y, Georgakopoulos D, Servillo G, Della Fazia MA, Kranias EG (2003-02-01). "Novel regulation of cardiac force-frequency relation by CREM (cAMP response element modulator)". FASEB Journal. 17 (2): 144–151. doi:10.1096/fj.01-0981com. ISSN 1530-6860. PMID 12554693. S2CID 17890025.
- ^ Sassone-Corsi P (2000-06-01). "CREM: a master-switch regulating the balance between differentiation and apoptosis in male germ cells". Molecular Reproduction and Development. 56 (2 Suppl): 228–229. doi:10.1002/(SICI)1098-2795(200006)56:2+<228::AID-MRD2>3.0.CO;2-B. ISSN 1040-452X. PMID 10824972. S2CID 21160055.
- ^ Sassone-Corsi P (1998-08-01). "CREM: a master-switch governing male germ cells differentiation and apoptosis". Seminars in Cell & Developmental Biology. 9 (4): 475–482. doi:10.1006/scdb.1998.0200. ISSN 1084-9521. PMID 9813195.
- ^ "CREM (cAMP responsive element modulator)". atlasgeneticsoncology.org. Retrieved 2016-10-16.
- ^ Fimia GM, De Cesare D, Sassone-Corsi P (Nov 2000). "A family of LIM-only transcriptional coactivators: tissue-specific expression and selective activation of CREB and CREM". Molecular and Cellular Biology. 20 (22): 8613–22. doi:10.1128/MCB.20.22.8613-8622.2000. PMC 102166. PMID 11046156.
- ^ Fimia GM, De Cesare D, Sassone-Corsi P (Mar 1999). "CBP-independent activation of CREM and CREB by the LIM-only protein ACT". Nature. 398 (6723): 165–9. Bibcode:1999Natur.398..165F. doi:10.1038/18237. PMID 10086359. S2CID 4424908.
- ^ Domschke K, Kuhlenbäumer G, Schirmacher A, Lorenzi C, Armengol L, DiBella D, Gratacos M, Garritsen HS, Nöthen MM (2003-02-01). "Human nuclear transcription factor gene CREM: genomic organization, mutation screening, and association analysis in panic disorder". American Journal of Medical Genetics Part B. 117B (1): 70–78. doi:10.1002/ajmg.b.10018. ISSN 1552-4841. PMID 12555239. S2CID 8884044.
- ^ Hamilton SP, Slager SL, Mayo D, Heiman GA, Klein DF, Hodge SE, Fyer AJ, Weissman MM, Knowles JA (2004-04-01). "Investigation of polymorphisms in the CREM gene in panic disorder". American Journal of Medical Genetics Part B. 126B (1): 111–115. doi:10.1002/ajmg.b.20121. ISSN 1552-4841. PMID 15048659. S2CID 43810849.
- ^ a b Krausz C, Sassone-Corsi P (2005-01-01). "Genetic control of spermiogenesis: insights from the CREM gene and implications for human infertility". Reproductive BioMedicine Online. 10 (1): 64–71. doi:10.1016/S1472-6483(10)60805-X. PMID 15705296.
- ^ Nantel F, Monaco L, Foulkes NS, Masquilier D, LeMeur M, Henriksén K, Dierich A, Parvinen M, Sassone-Corsi P (1996-03-14). "Spermiogenesis deficiency and germ-cell apoptosis in CREM-mutant mice". Nature. 380 (6570): 159–162. Bibcode:1996Natur.380..159N. doi:10.1038/380159a0. ISSN 0028-0836. PMID 8600390. S2CID 4278745.
- ^ Jasem E, Nasim J, Gholamreza M, Naser S, Nader M, Maryam SL, Abbas N, Vahid R (2010-10-01). "Evaluation of the effects of Salvia hypoleuca on the cAMP-responsive element modulator (CREM) gene expression and spermatogenesis in rat". Middle East Fertility Society Journal. 15 (4): 274–277. doi:10.1016/j.mefs.2010.08.002.
- ^ Mazaheri M, Shahdadi V, Nazari Boron A (2016-10-16). "Molecullar and biochemical effect of alcoholic extract of Alpinia galanga on rat spermatogenesis process". Iranian Journal of Reproductive Medicine. 12 (11): 765–770. ISSN 1680-6433. PMC 4330656. PMID 25709632.
- ^ Juang YT, Wang Y, Solomou EE, Li Y, Mawrin C, Tenbrock K, Kyttaris VC, Tsokos GC (2005-04-01). "Systemic lupus erythematosus serum IgG increases CREM binding to the IL-2 promoter and suppresses IL-2 production through CaMKIV". Journal of Clinical Investigation. 115 (4): 996–1005. doi:10.1172/JCI22854. ISSN 0021-9738. PMC 1070410. PMID 15841182.
Further reading
- Don J, Stelzer G (Feb 2002). "The expanding family of CREB/CREM transcription factors that are involved with spermatogenesis". Molecular and Cellular Endocrinology. 187 (1–2): 115–24. doi:10.1016/S0303-7207(01)00696-7. PMID 11988318. S2CID 23023082.
- Yan C, Miller CL, Abe J (Mar 2007). "Regulation of phosphodiesterase 3 and inducible cAMP early repressor in the heart". Circulation Research. 100 (4): 489–501. doi:10.1161/01.RES.0000258451.44949.d7. PMC 4115784. PMID 17332439.
- Pongubala JM, Atchison ML (Apr 1995). "Activating transcription factor 1 and cyclic AMP response element modulator can modulate the activity of the immunoglobulin kappa 3' enhancer". The Journal of Biological Chemistry. 270 (17): 10304–13. doi:10.1074/jbc.270.17.10304. PMID 7730336.
- Walker WH, Sanborn BM, Habener JF (Dec 1994). "An isoform of transcription factor CREM expressed during spermatogenesis lacks the phosphorylation domain and represses cAMP-induced transcription". Proceedings of the National Academy of Sciences of the United States of America. 91 (26): 12423–7. Bibcode:1994PNAS...9112423W. doi:10.1073/pnas.91.26.12423. PMC 45450. PMID 7809053.
- Fujimoto T, Fujisawa J, Yoshida M (Feb 1994). "Novel isoforms of human cyclic AMP-responsive element modulator (hCREM) mRNA". Journal of Biochemistry. 115 (2): 298–303. doi:10.1093/oxfordjournals.jbchem.a124332. PMID 8206879.
- de Groot RP, den Hertog J, Vandenheede JR, Goris J, Sassone-Corsi P (Oct 1993). "Multiple and cooperative phosphorylation events regulate the CREM activator function". The EMBO Journal. 12 (10): 3903–11. doi:10.1002/j.1460-2075.1993.tb06068.x. PMC 413673. PMID 8404858.
- Bodor J, Walker W, Flemington E, Spetz AL, Habener JF (Dec 1995). "Modulation of Tax and PKA-mediated expression of HTLV-I promoter via cAMP response element binding and modulator proteins CREB and CREM". FEBS Letters. 377 (3): 413–8. doi:10.1016/0014-5793(95)01299-0. PMID 8549766. S2CID 45422227.
- Nantel F, Monaco L, Foulkes NS, Masquilier D, LeMeur M, Henriksén K, Dierich A, Parvinen M, Sassone-Corsi P (Mar 1996). "Spermiogenesis deficiency and germ-cell apoptosis in CREM-mutant mice". Nature. 380 (6570): 159–62. Bibcode:1996Natur.380..159N. doi:10.1038/380159a0. PMID 8600390. S2CID 4278745.
- Blendy JA, Kaestner KH, Weinbauer GF, Nieschlag E, Schütz G (Mar 1996). "Severe impairment of spermatogenesis in mice lacking the CREM gene". Nature. 380 (6570): 162–5. Bibcode:1996Natur.380..162B. doi:10.1038/380162a0. PMID 8600391. S2CID 4337719.
- Bodor J, Spetz AL, Strominger JL, Habener JF (Apr 1996). "cAMP inducibility of transcriptional repressor ICER in developing and mature human T lymphocytes". Proceedings of the National Academy of Sciences of the United States of America. 93 (8): 3536–41. Bibcode:1996PNAS...93.3536B. doi:10.1073/pnas.93.8.3536. PMC 39645. PMID 8622971.
- Gellersen B, Kempf R, Telgmann R (Jan 1997). "Human endometrial stromal cells express novel isoforms of the transcriptional modulator CREM and up-regulate ICER in the course of decidualization". Molecular Endocrinology. 11 (1): 97–113. doi:10.1210/mend.11.1.9875. PMID 8994192.
- Laurance ME, Kwok RP, Huang MS, Richards JP, Lundblad JR, Goodman RH (Jan 1997). "Differential activation of viral and cellular promoters by human T-cell lymphotropic virus-1 tax and cAMP-responsive element modulator isoforms". The Journal of Biological Chemistry. 272 (5): 2646–51. doi:10.1074/jbc.272.5.2646. PMID 9006899.
- Bonny C, Cooker LA, Goldberg E (Mar 1998). "Deoxyribonucleic acid-protein interactions and expression of the human testis-specific lactate dehydrogenase promoter: transcription factor Sp1 plays a major role". Biology of Reproduction. 58 (3): 754–9. doi:10.1095/biolreprod58.3.754. PMID 9510963.
- Müller FU, Bokník P, Knapp J, Neumann J, Vahlensieck U, Oetjen E, Scheld HH, Schmitz W (Sep 1998). "Identification and expression of a novel isoform of cAMP response element modulator in the human heart". FASEB Journal. 12 (12): 1191–9. doi:10.1096/fasebj.12.12.1191. PMID 9737722. S2CID 34554530.
- Fimia GM, De Cesare D, Sassone-Corsi P (Mar 1999). "CBP-independent activation of CREM and CREB by the LIM-only protein ACT". Nature. 398 (6723): 165–9. Bibcode:1999Natur.398..165F. doi:10.1038/18237. PMID 10086359. S2CID 4424908.
- Pati D, Meistrich ML, Plon SE (Jul 1999). "Human Cdc34 and Rad6B ubiquitin-conjugating enzymes target repressors of cyclic AMP-induced transcription for proteolysis". Molecular and Cellular Biology. 19 (7): 5001–13. doi:10.1128/mcb.19.7.5001. PMC 84326. PMID 10373550.
- Inada A, Someya Y, Yamada Y, Ihara Y, Kubota A, Ban N, Watanabe R, Tsuda K, Seino Y (Jul 1999). "The cyclic AMP response element modulator family regulates the insulin gene transcription by interacting with transcription factor IID". The Journal of Biological Chemistry. 274 (30): 21095–103. doi:10.1074/jbc.274.30.21095. PMID 10409662.
- Zauli G, Secchiero P, Rodella L, Gibellini D, Mirandola P, Mazzoni M, Milani D, Dowd DR, Capitani S, Vitale M (Feb 2000). "HIV-1 Tat-mediated inhibition of the tyrosine hydroxylase gene expression in dopaminergic neuronal cells". The Journal of Biological Chemistry. 275 (6): 4159–65. doi:10.1074/jbc.275.6.4159. PMID 10660577.
External links
- CREM+protein,+human at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
- Human CREM genome location and CREM gene details page in the UCSC Genome Browser.
- v
- t
- e
-
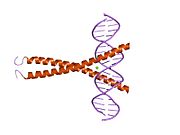 1dh3: CRYSTAL STRUCTURE OF A CREB BZIP-CRE COMPLEX REVEALS THE BASIS FOR CREB FAIMLY SELECTIVE DIMERIZATION AND DNA BINDING
1dh3: CRYSTAL STRUCTURE OF A CREB BZIP-CRE COMPLEX REVEALS THE BASIS FOR CREB FAIMLY SELECTIVE DIMERIZATION AND DNA BINDING
This article incorporates text from the United States National Library of Medicine, which is in the public domain.